GATK Joint Genotyping#
23/03/2022
For germline short variants (SNPs and indels), the GATK workflow includes a joint analysis step that empowers variant discovery by providing the ability to leverage population-wide information from a cohort of multiple samples, allowing us to detect variants with great sensitivity and genotype samples as accurately as possible. Atgenomix demonstrates the power of the SeqsLab platform in this case study by executing the GATK Joint Genotyping pipeline, which is based on the Github Project, JointGenotyping_v1.5.1 ( ).
).
Main WDL#
The main WDL used in this example can be referenced from the Atgenomix public GitHub account ( ).
).
WOM graph#
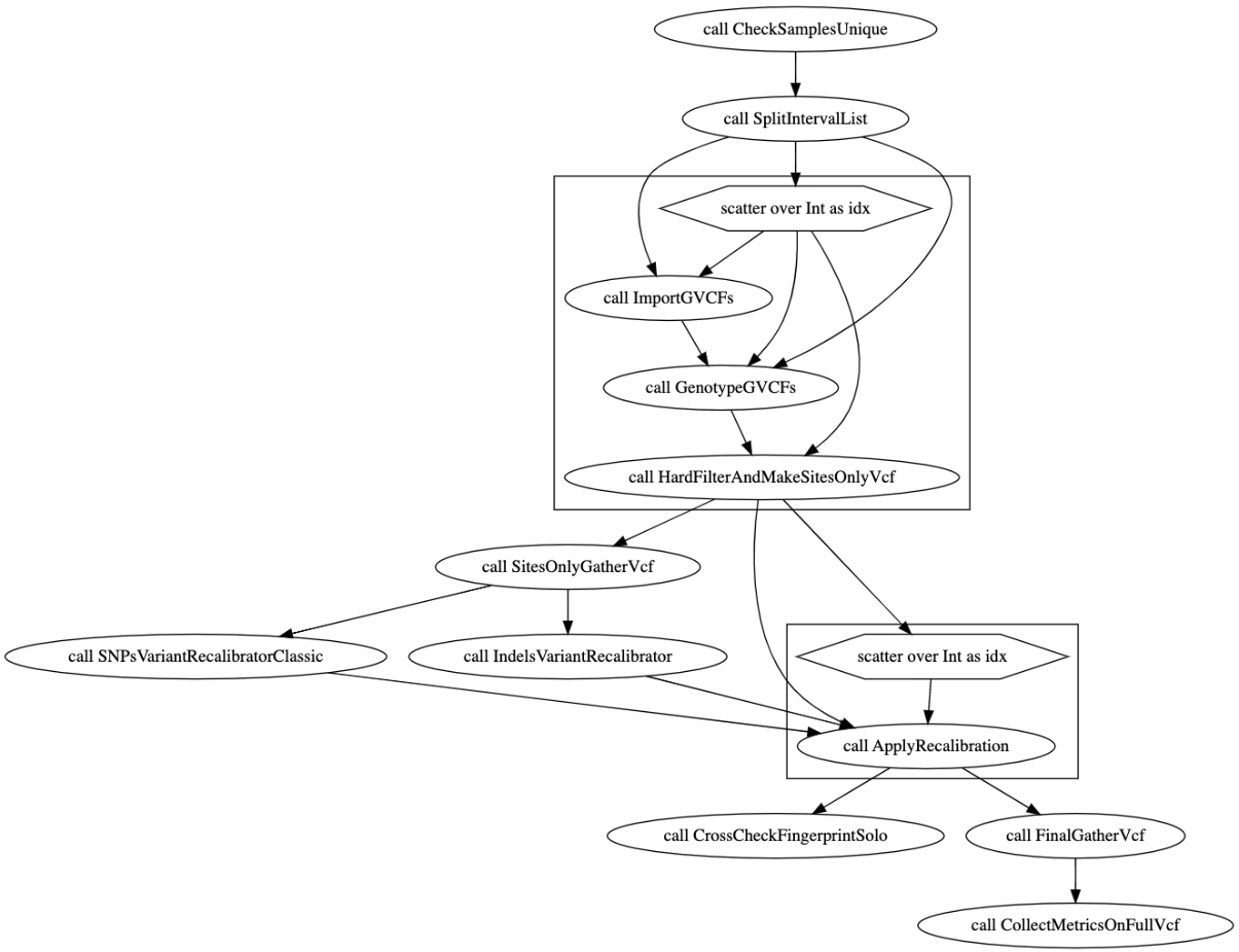
Performance Evaluation#
Performance (Time) |
52 samples |
60 samples |
|---|---|---|
CheckSamplesUnique |
3m:50s |
|
SplitIntervalList |
26s |
|
ImportGVCFs |
24m:31s |
|
GenotypeGVCFs |
12m:25s |
|
HardFilterAndMakeSitesOnlyVcf |
1m:31s |
|
SitesOnlyGatherVcf |
2m:33s |
|
IndelsVariantRecalibrator |
54m:20s |
|
SNPsVariantRecalibratorClassic |
1h:18m:45s |
|
ApplyRecalibration |
2m:19s |
|
CrossCheckFingerprintSolo |
5h:23m:00s |
|
FinalGatherVcf |
9m:56s |
|
CollectMetricsOnFullVcf |
1h:11m:49s |
|
Total |
7h:43m:21s |
