Configure the execution file#
After developing a tool, the WDL directory (containing all imported OpenWDL files), inputs.json file, and registered Docker runtime images should be ready. At this point, you can proceed to the next step of the tool onboarding process, which is preparing the SeqsLab execs.json file.
The following diagram provides an overview of the entire process, which includes several manual steps.
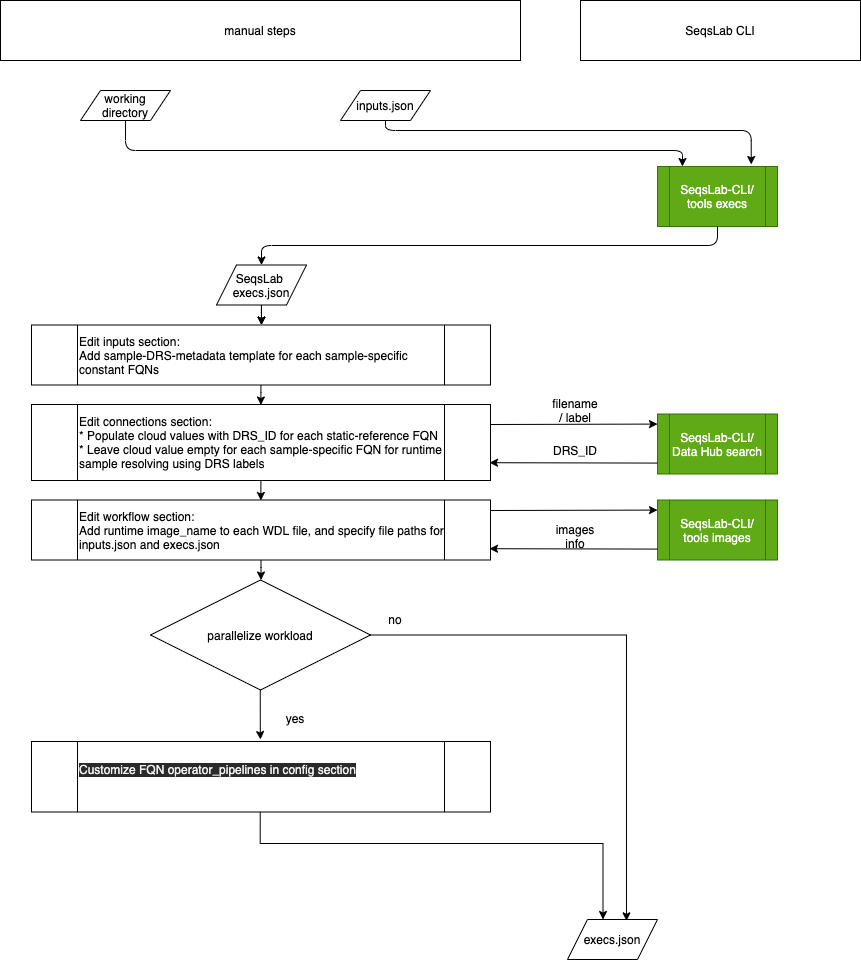
Create the execs.json file#
The first step is to generate an execs.json template using the SeqsLab CLI tools execs command.
seqslab tools execs \
--working-dir /home/ubuntu/seqslab_workflows/src/ \
--inputs inputs/inputs_germline-gatk4-snpindel_hg38.json \
--main-wdl wdl/germline-gatk4-snpindel.wdl \
--output execs/germline-gatk4-snpindel.json
The execs.json file is extended from inputs.json. In addition to the inputs section, the execs.json file contains the connections, workflows, calls, configs, and operator_pipelines sections. Running the tools execs command
generates a template for execs.json containing the information from the inputs.json file, default SeqsLab configuration settings, and the additional sections which you will need to manually complete.
Inputs section#
Sample-specific FQNs / static-reference FQNs#
The inputs section includes the mapping of fully qualified names (FQNs) of a WDL workflow to their corresponding values. The FQNs can be categorized as either a static-reference group, which remains unchanged for all samples run on this workflow, or as a sample-specific group, which is assigned in a sample-by-sample manner.
The following example uses the GATK4Fq2Gvcf workflow, where GATK4Fq2Gvcf.refFasta, GATK4Fq2Gvcf.dbSNPVcf, and GATK4Fq2Gvcf.knownIndelsSitesVCFs all belong to the
static-reference group since they are static reference genome files. Meanwhile, GATK4Fq2Gvcf.fastqFiles and GATK4Fq2Gvcf.sampleName both belong to the sample-specific group.
{
# sample-specific FQNs
"GATK4Fq2Gvcf.fastqFiles": [
"/mnt/reads/NA12878_r1.fq.gz",
"/mnt/reads/NA12878_r2.fq.gz"
],
"GATK4Fq2Gvcf.sampleName": "NA12878",
# static-reference FQNs
"GATK4Fq2Gvcf.refFasta": "hg38/Homo_sapiens_assembly38.fasta",
"GATK4Fq2Gvcf.refFastaIndex": "hg38/Homo_sapiens_assembly38.fasta.fai",
"GATK4Fq2Gvcf.refBwt": "hg38/Homo_sapiens_assembly38.fasta.64.bwt",
"GATK4Fq2Gvcf.refDict": "hg38/Homo_sapiens_assembly38.dict",
"GATK4Fq2Gvcf.gatkPath": "/gatk/gatk-4.2.0.0/gatk",
...
}
It is important to differentiate the two types of FQNs because it affects how a tool is registered on the SeqsLab TRS. For the static-reference group, the FQNs remain constant. Meanwhile, for the sample-specific group, the FQNs change according to each given sample file, which is presumably registered as a Data Repository Service (DRS) object.
Sample-DRS-metadata template#
SeqsLab platform provides a sample-DRS-metadata template syntax, in the format of ~{FQN:metadata.key}, to render constant
sample-specific FQN values based on the DRS metadata of a specific FQN. DRS metadata are registered during the sample files upload and registration process based on the Run Sheet information.
In the previous example, we configured the FQN GATK4Fq2Gvcf.sampleName as "~{GATK4Fq2Gvcf.fastqFiles:sample.Sample_ID}", indicating that SeqsLab should render GATK4Fq2Gvcf.sampleName based on the metadata sample.Sample_ID of the DRS object assigned to the FQN of GATK4Fq2Gvcf.fastqFiles. The DRS metadata is attached in the sample data upload and register process, which is managed with the datahub upload-runsheet command information. As such, the constant FQN GATK4Fq2Gvcf.sampleName can be altered along with the FQN GATK4Fq2Gvcf.fastqFiles, and the TRS tool configured accordingly can be used with many different samples.
{
# sample-specific FQNs
"GATK4Fq2Gvcf.fastqFiles": [
"/mnt/reads/NA12878_r1.fq.gz",
"/mnt/reads/NA12878_r2.fq.gz"
],
"GATK4Fq2Gvcf.sampleName": "~{GATK4Fq2Gvcf.fastqFiles:sample.Sample_ID}",
# static-reference FQNs
"GATK4Fq2Gvcf.refFasta": "hg38/Homo_sapiens_assembly38.fasta",
"GATK4Fq2Gvcf.refFastaIndex": "hg38/Homo_sapiens_assembly38.fasta.fai",
"GATK4Fq2Gvcf.refBwt": "hg38/Homo_sapiens_assembly38.fasta.64.bwt",
"GATK4Fq2Gvcf.refDict": "hg38/Homo_sapiens_assembly38.dict",
"GATK4Fq2Gvcf.gatkPath": "/gatk/gatk-4.2.0.0/gatk",
...
}
Connections section#
DRS_ID for file FQNs#
The connections section provides a mapping of each WDL file-typed FQN to its local and cloud paths. The file-typed FQNs can also be separated into a sample-specific FQN group and a static-reference FQN group. The execs.json template has the FQNs and the local paths filled based on the inputs.json file, but the cloud paths are left blank, as shown in the following example.
"connections": [
# sample-specific FQNs
{
"fqn": "GATK4Fq2Gvcf.fastqFiles",
"local": [
"/mnt/reads/NA12878_r1.fq.gz",
"/mnt/reads/NA12878_r2.fq.gz"
],
"cloud": []
},
# static-reference FQNs
{
"fqn": "GATK4Fq2Gvcf.refFasta",
"local": [
"hg38/Homo_sapiens_assembly38.fasta"
],
"cloud": []
},
{
"fqn": "GATK4Fq2Gvcf.refFastaIndex",
"local": [
"hg38/Homo_sapiens_assembly38.fasta.fai"
],
"cloud": []
},
{
"fqn": "GATK4Fq2Gvcf.refSa",
"local": [
"hg38/Homo_sapiens_assembly38.fasta.64.sa"
],
"cloud": []
},
...
]
Static-reference FQNs#
For each static-reference FQNs, you will need to fill in the DRS URI in the cloud section, so as to identify which DRS object will actually be used when the tool is executed on the SeqsLab platform.
Atgenomix recommends two methods for retrieving the DRS URI. The first method uses the datahub search command that can take either the DRS object tag or DRS object name as a query parameter to find the corresponding DRS URI.
seqslab datahub search --name Homo_sapiens_assembly38.fasta
seqslab datahub search --tag hg38/Homo_sapiens_assembly38-fasta
Below is an example output for the above command:
{
"objects": [
{
"self_uri": "drs://api.seqslab.net/drs_010MAvDKw23Y5yb",
"name": "Homo_sapiens_assembly38.fasta",
"id": "hg38_Homo_sapiens_assembly38-fasta",
"tags": [
"hg38/Homo_sapiens_assembly38.fasta"
]
}
]
}
The second method makes use of custom DRS IDs and tags to simplify the query process. As described in the Customizing the DRS metadata section, you can create the customized DRS ID hg38_Homo_sapiens_assembly38-fasta based on a simple conversion rule using the local path information (hg38/Homo_sapiens_assembly38.fasta). As such, the DRS URI of the corresponding DRS object can be directly inferred from the hostname (drs://api.seqslab.net/) and the local path.
Sample-specific FQNs#
For the sample-specific FQNs, on the other hand, Atgenomix recommends leaving the cloud section empty, so that the SeqsLab Workflow Execution Service (WES) runtime DRS object resolving mechanism will take effect. The mechanism resolves the DRS object at runtime based on the DRS object tags, which are specified in the Run_Name, Read1_Tag, and Read2_Tag columns of the Run Sheet.
"connections": [
# sample-specific FQNs, leave it blank for WES runtime DRS to resolve
{
"fqn": "GATK4Fq2Gvcf.fastqFiles",
"local": [
"/mnt/reads/NA12878_r1.fq.gz",
"/mnt/reads/NA12878_r2.fq.gz"
],
"cloud": []
},
# static-reference FQNs, fill DRS ID for the cloud section of each FQN
{
"fqn": "GATK4Fq2Gvcf.refFasta",
"local": [
"hg38/Homo_sapiens_assembly38.fasta"
],
"cloud": ["drs://api.seqslab.net/drs_010MAvDKw23Y5yb"]
},
{
"fqn": "GATK4Fq2Gvcf.refFastaIndex",
"local": [
"hg38/Homo_sapiens_assembly38.fasta.fai"
],
"cloud": ["drs://api.seqslab.net/drs_bIW4jKMob4tEijO"]
},
{
"fqn": "GATK4Fq2Gvcf.refSa",
"local": [
"hg38/Homo_sapiens_assembly38.fasta.64.sa"
],
"cloud": ["drs://api.seqslab.net/drs_A25LXPutuqxiYHt"]
},
...
]
Workflow section#
The workflow section provides a list of files that will be registered in the TRS object, where all WDL files, the inputs.json file, and the execs.json file should be included. For each of the WDL files, the execs.json template has the file_type, path, and name properties filled out.
You will need to provide the Docker runtime images information. You will also need to replace the instances of “inputs.json” and “execs.json” in the template with their relative paths in the working directory.
"workflow": [
{
"name": "e2e-gatk4-germline-snp-indels.wdl",
"path": "e2e-workflows/atgx/e2e-gatk4-germline-snp-indels.wdl",
"file_type": "PRIMARY_DESCRIPTOR",
"image_name": ""
},
{
"name": "processing-for-variant-discovery-gatk4.wdl",
"path": "gatk4-data-processing/processing-for-variant-discovery-gatk4.wdl",
"file_type": "SECONDARY_DESCRIPTOR",
"image_name": ""
},
{
"name": "haplotypecaller-gvcf-gatk4.wdl",
"path": "gatk4-germline-snps-indels/haplotypecaller-gvcf-gatk4.wdl",
"file_type": "SECONDARY_DESCRIPTOR",
"image_name": ""
},
{
"path": "inputs.json",
"file_type": "TEST_FILE"
},
{
"path": "exec.json",
"file_type": "EXECUTION_FILE"
}
],
The following is an example of a completed workflow section:
"workflow": [
{
"name": "e2e-gatk4-germline-snp-indels.wdl",
"path": "e2e-workflows/atgx/e2e-gatk4-germline-snp-indels.wdl",
"file_type": "PRIMARY_DESCRIPTOR",
"image_name": "germline-gatk4-snpindel-1.0_ubuntu-20.04:2022-03-01-01-03"
},
{
"name": "processing-for-variant-discovery-gatk4.wdl",
"path": "gatk4-data-processing/processing-for-variant-discovery-gatk4.wdl",
"file_type": "SECONDARY_DESCRIPTOR",
"image_name": "germline-gatk4-snpindel-1.0_ubuntu-20.04:2022-03-01-01-03"
},
{
"name": "haplotypecaller-gvcf-gatk4.wdl",
"path": "gatk4-germline-snps-indels/haplotypecaller-gvcf-gatk4.wdl",
"file_type": "SECONDARY_DESCRIPTOR",
"image_name": "germline-gatk4-snpindel-1.0_ubuntu-20.04:2022-03-01-01-03"
},
{
"path": "inputs/hg38-e2e-gatk4-germline-snp-indels.json",
"file_type": "TEST_FILE"
},
{
"path": "execs/hg38-e2e-gatk4-germline-snp-indels.json",
"file_type": "EXECUTION_FILE"
}
],
Config section#
The config section provides a full list of file-typed internal FQNs of the workflows, and their corresponding operator_pipeline settings. By default, all file-typed internal FQNs will be assigned to the default operator_pipeline setting opp_generic-singular_auto, which does not apply any data parallelization scheme. As such, for a tool designed to be run without parallel execution enhancement, the config section can be left as is. However, for tools that require data parallelization, additional steps are required. For details, see Pipeline operators.
